AdenoZAP Kit
Faster method for constructing replication-deficient adenovirus vectors containing transgenes in the E1 region, only one cloning step in E. coli
- Transgene insertion in E1 region
- E3 region: WT or 2.7 kb deletion
- E4 region: WT or 1.2 kb deletion
- Fiber: WT Ad5 or hybrid Ad5/35
- Maximum cargo capacity: 9.6 kb
- Kit contains: E1 shuttle plasmid, RightZAP DNA, adenovirus DNA used as positive control for virus rescue
Make your kit
RightZap1.1 DNA Visit Product Page »
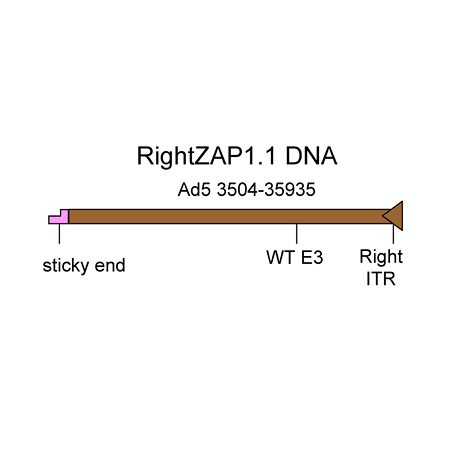
The RightZAP1.1 DNA is a 32.4 kb linear DNA fragment that encompasses bp 3504-right end of the Ad5 genome. The E3 region is intact (WT). The DNA was purified from a cosmid. It is used in combination with the shuttle vectors pZAP1.1 or pZAP1.2 to construct replication-deficient adenoviruses containing transgenes in place of the E1 region of the Ad5 genome. The maximum transgene capacity of the resulting virus is 5.2 kb.
pZAP1.1 Visit Product Page »
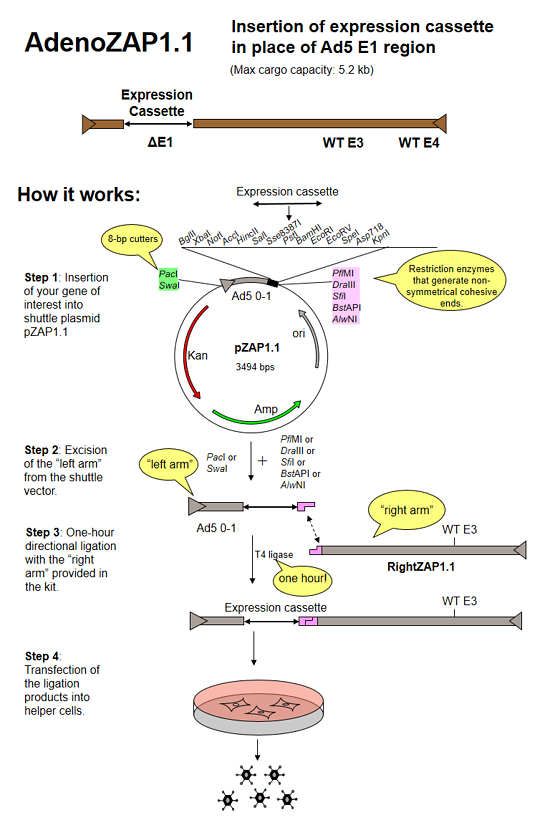
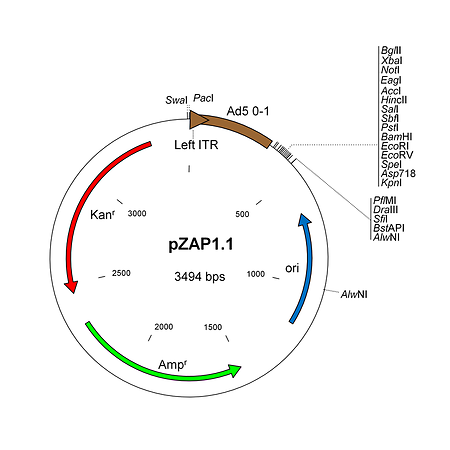
pZAP1.1 is a 3.5 kb shuttle plasmid designed for inserting expression cassettes in place of the E1 region of the Ad5 genome, in combination with RightZAP1.1 (WT E3), RightZAP1.2 (ΔE3), RightZAP1.3 (ΔE3 + Ad5/35 knob) and other vectors from the AdenoZAP1.0 cloning system. It contains a multiple cloning site downstream from the left ITR and packaging signal of the Ad5 genome. Expression cassettes inserted into this site should contain a promoter, cDNA, and polyA signal. The left arm DNA (which corresponds to the left ITR, packaging signal and expression cassette) can be excised from the vector with either PacI or SwaI on one side, and either PflMI, DraIII, SfiI, BstAPI or AlwNI on the other side.
There are two ways to generate adenovirus vectors with the AdenoZAP system:
1. In vitro ligation and transfection
The AdenoZAP™ cloning system is primarily based on the in vitro ligation of two DNA molecules: the "left arm" and the "right arm".
The left arm is obtained after cloning your gene of interest into a pZAP shuttle plasmid, digesting the resulting plasmid with restriction enzymes and purifying on agarose gel the DNA fragment containing the left ITR, packaging signal and your gene of interest.
The right arm is provided in the kit. It contains most of the adenovirus genome (from bp 3504 in WT Ad5 to the right end, with several options regarding the status of the E3 and E4 regions, and fiber gene). It originates from a bacterial cosmid rather than from virions, thereby eliminating the danger for a possible viral contamination. It will generate virus only after being ligated to the left arm that contains your expression cassette and the packaging signal.
We have designed the AdenoZAP™ cloning system in such a way that the ligation of both left and right arms is directional. Because of the non-symmetrical nature of the sticky ends generated by excising the left arm from the shuttle plasmid, the ligation of the left and right arms should generate only one type of viral DNA molecule. Consequently, a homogeneous virus population should be generated upon transfection of the helper cells. In addition, such ligation is performed to almost completion in less than an hour using an ordinary T4 DNA ligase.
The ligation product is directly transfected into a helper cell line such as 293 cells. In general, viral plaques appear several days after the transfection.
2. Transfection and in vivo recombination
The second way of creating recombinant adenovirus vectors with the AdenoZAP system is via homologous DNA recombination in helper cells such as 293 cells. The expression cassettes are cloned into shuttle plasmids pZAP1.2 or pZAP2.2, which contain a 400 bp-long Ad5 DNA sequence downstream from the multiple cloning site. This Ad5 DNA sequence is part of the pIX gene (bp 3504-3907 in the WT Ad5 genome), and is identical to the left end of the RightZAP DNAs. The shuttle plasmid is linearized with either PacI or SwaI (whichever is not present in your gene of interest), then transfected into 293 cells together with a RightZAP DNA. Since both DNA molecules share identical sequences, they can be joined by homologous recombination in 293 cells to create functional recombinant adenovirus vectors. This approach offers two advantages: (1) the shuttle plasmid needs to be digested with only one restriction enzyme (PacI or SwaI) and (2) there is no need to purify a DNA fragment on agarose gel. However, the shuttle plasmid must be transfection-grade DNA.
Choosing the AdenoZAP Cloning Kit Suitable for your application.
The AdenoZAP Cloning System exists in different versions and each version can be used in different ways. The following table will help you decide which one is the most appropriate to your application.
| Kit | Shuttle Plasmid | Enzymes available for Left Arm Excision | Right Arm | E3 | Fiber | E4 | Max Cargo | |
|---|---|---|---|---|---|---|---|---|
| Left End | Right End | |||||||
| AdenoZAP1.1 | pZAP1.1 or pZAP1.2 | Pac I, Swa I | Sfi I, BstAPI, DraIII, AlwNI, PflMI | RightZAP1.1 | WT | Ad5 | WT | 5.2 kb |
| AdenoZAP1.2 | pZAP1.1 or pZAP1.2 | Pac I, Swa I | Sfi I, BstAPI, DraIII, AlwNI, Pfl MI | RightZAP1.2 | Δ 2.7 kb | Ad5 | WT | 7.9 kb |
| AdenoZAP1.3 | pZAP1.1 or pZAP1.2 | Pac I, Swa I | Sfi I, BstAPI, DraIII, AlwNI, Pfl MI | RightZAP1.3 | Δ 2.7 kb | Ad5/35 | WT | 8.3 kb |
| AdenoZAP1.4 | pZAP1.1 or pZAP1.2 | Pac I, Swa I | Sfi I, BstAPI, DraIII, AlwNI, Pfl MI | RightZAP1.4 | Δ 2.7 kb | Ad5/35 | Δ 1.2 kb | 9.6 kb |
| AdenoZAP2.1 | pZAP2.1 or pZAP2.2 | Pac I, Swa I | I-SceI | RightZAP2.1 | WT | Ad5 | WT | 5.2 kb |
| AdenoZAP2.2 | pZAP2.1 or pZAP2.2 | Pac I, Swa I | I-SceI | RightZAP2.2 | Δ 2.7 kb | Ad5 | WT | 7.9 kb |
- Only one cloning step in E. coli
The only cloning you have to perform in E. coli is inserting your expression cassette into a small shuttle vector.
- No large plasmid (or cosmid) construction and purification
Constructing large plasmids or cosmids is not obvious for inexperienced researchers. In addition, purifying them in a quality suitable for transfection (i.e. without E. coli chromosomal DNA) is difficult using column chromatography and is better performed on CsCl gradients, which is time-consuming and hazardous.
- No plasmid midi-prep or maxi-prep necessary
Although using a purified preparation of your shuttle plasmid maximizes the chances of getting your recombinant virus, it is not necessary. You can use mini-prep DNA: just clean it up after verifying the identity of your clone, digest it with the appropriate enzymes and purify the fragment of interest on agarose gel. A few micrograms plasmid DNA are usually sufficient to perform the whole procedure.
- A versatile set of endonucleases available to generate the left arm
Two versions of AdenoZAP cloning system are available. In AdenoZAP1.0, two 8-base cutters (PacI and SwaI) are available to digest your shuttle vector next to the left ITR, and four 6-base cutters (AlwNI,
BstAPI, DraIII, PflMI) and one 8-base cutter (SfiI) are available to cut your shuttle vector downstream from your expression cassette. In AdenoZAP2.0, the same 8-base cutters (PacI and SwaI) are available to digest your shuttle vector next to the left ITR, while an 18-base intron-encoded endonuclease (I-SceI) is available to cut the shuttle vector downstream from your expression cassette. It is therefore likely that this system will be useful in a very large number of applications.
- Fast
With only one cloning step in E. coli, a DNA digestion, gel purification and a 1-hour ligation before the transfection can be performed, the overall hands-on time is strongly reduced compared to most of the other methods currently available. If well organized, one can perform the DNA transfection into helper cells only 3 days after starting the cloning of an expression cassette into the shuttle vector and harvest the first virus plaques 3-7 days later.
Adenovirus Expression Vectors
The AdenoZAP cloning system is ideal for constructing replication-deficient adenovirus expression vectors, wherein a transgene expression cassette (promoter + coding sequence + polyA signal) is inserted in place of the E1 region. This is the most popular type of adenovirus vector, also called "first-generation". The applications include:
- Transient gene expression in a large variety of cell types and tissues
- Gene therapy vectors
- Vaccine vectors
The AdenoZAP cloning system can also be used to construct multi-cistronic adenovirus vectors expressing two or more transgenes. This can be achieved in multiple ways: (1) by inserting two or more expression cassettes next to each other in the E1 region (pZAP shuttle plasmid); (2) by using IRES-based dicistronic transcriptional cassettes, or (3) by making fusion proteins with self-cleaving P2A peptides. Another option is to use the AdenoQuick1.0 or AdenoQuick2.0 systems, which allow for inserting expression cassettes at various locations in the adenovirus genome.
- Ad5 backbone
- E1 region:
- 3.1 kb deletion, corresponding to bp 354-3,503 in the Ad5 genome
- E3 region:
- WT
- 2.7 kb BglII deletion, corresponding to bp 28,133-30,818 in the Ad5 genome
- Fiber:
- WT
- Hybrid Ad5/35
- E4 region:
- WT
- 1.2 kb deletion, corresponding to bp 35,319-35,355 in the Ad5 genome and including ORF1-4
AdenoZAP Manual
The AdenoZAP Manual contains all the instructions for constructing adenovirus expression vectors using the AdenoZAP system, including:
- Shuttle plasmid construction
- Virus Rescue
The manual contains also safety guidelines for handling adenovirus vectors, detailed plasmid maps, and troubleshooting guide.